
I’d like to highlight a recent publication entitled: Quantitative determination of SLC2A1 variant functional effects in GLUT1 deficiency syndrome by Dr. Naeimeh Tayebi. Dr. Tayebi is part of Dr. Christina Gurnett’s lab in the Department of Neurology at Washington University School of Medicine in St. Louis. Dr. Gurnett was our Million Dollar Bike Ride grantee in 2021 and this publication is the result of her team’s work from this grant.
The reason behind this study is that many patients with Glut1 Deficiency, now more than ever, are being diagnosed by genetic testing, however, there are now about 300 variants of uncertain significance (VUS). VUS are variants or mutations on a gene that are not clearly connected to a health condition. In many cases these variants are so rare in the population that little information is known about them. Therefore, in some cases, to give a definite diagnosis of Glut1 Deficiency through genetic testing, when there are so many VUS, is more difficult.
The hypothesis for this study was that because SLC2A1, the gene responsible for Glut1 Deficiency, is required for growth of the cell line HAP1, the introduction of single nucleotide variants (SVNs) into the single copy of the SLC2A1 gene in these cells would allow them to determine which variants are detrimental for GLUT1 to function. SNVs occur when a single nucleotide [adenine (A), thymine (T), guanine (G) or cytosine (C)] is changed in the genome sequence.
The objective of this study was to demonstrate the use of a growth assay to quantify the impact on the function of single nucleotide variants (SNVs) in the SLC2A1 gene.
The methods for this study were to first introduce SNVs into the non-mutated SLC2A1 gene using a special molecular technique called CRISPR-Cas9. This technique enables researchers to edit parts of the genome by removing, adding or altering sections of the DNA sequence. Through different molecular biology techniques, Dr. Gurnett and her team produced 40 SNV of the SLC2A1 gene located in exons 2 and 3 of the gene as seen in the diagram below:
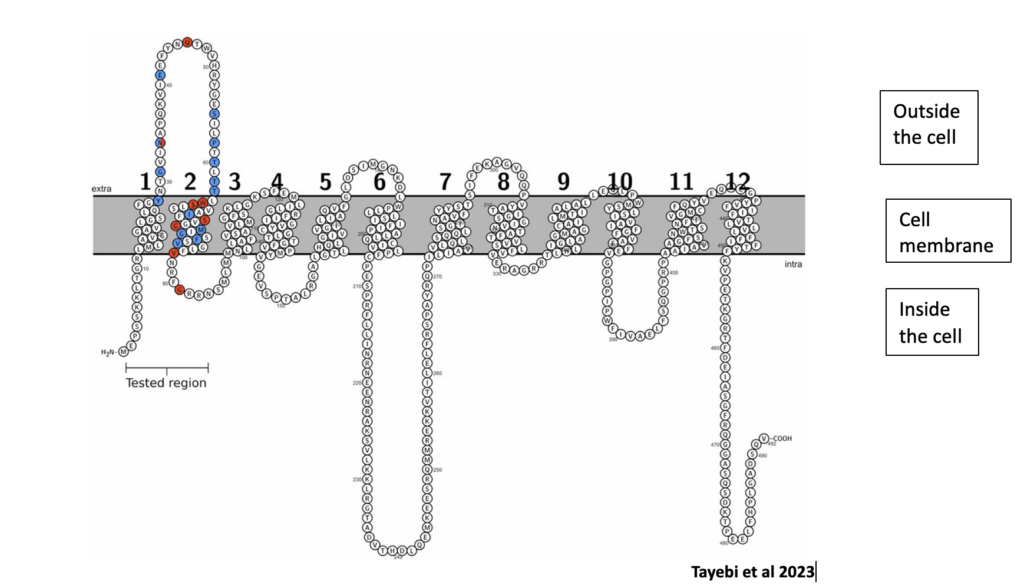
Diagram of the GLUT1 protein showing the locations of variants from exon 2 and 3. Variants identified in patients with clinical data are highlighted in red, others are in blue. The circled letters represent amino acids or the building blocks of proteins. The numbers 1-12 represent the protein’s transmembrane domains. Transmembrane domains of a protein are functional regions that span the cell membrane.
The results show that among the 40 SNVs there were 6 variants reported as causing Glut1 Deficiency (pathogenic), 7 not causing disease (benign) and 7 VUS. The remaining variants had not been reported before.
A growth assay was performed for the HAP1 cells containing the SNVs to analyze the functional effect of the 40 different SNVs in SLC2A1, using enough replicas or reproductions to ensure the results would be statistically significant. In addition, HAP1 cells without any type of Single Nucleotide Variant were evaluated in the same growth assay to compare with the HAP1 cells containing the variants. Each cell line was scored for their growth. High negative scores indicate a detrimental effect on the function of the protein, and were scored by nonsense variants. Nonsense variants are variants that lead to a premature stop of the protein being made.
On the other hand, low negative scores or positive scores indicated a mild effect or no effect on the protein function and were scored by missense variants or synonymous variants. Missense variants are variants that lead to a protein with a change in the amino acid different from the normal protein. A synonymous variant is a variant that does not change the amino acid from the normal protein.
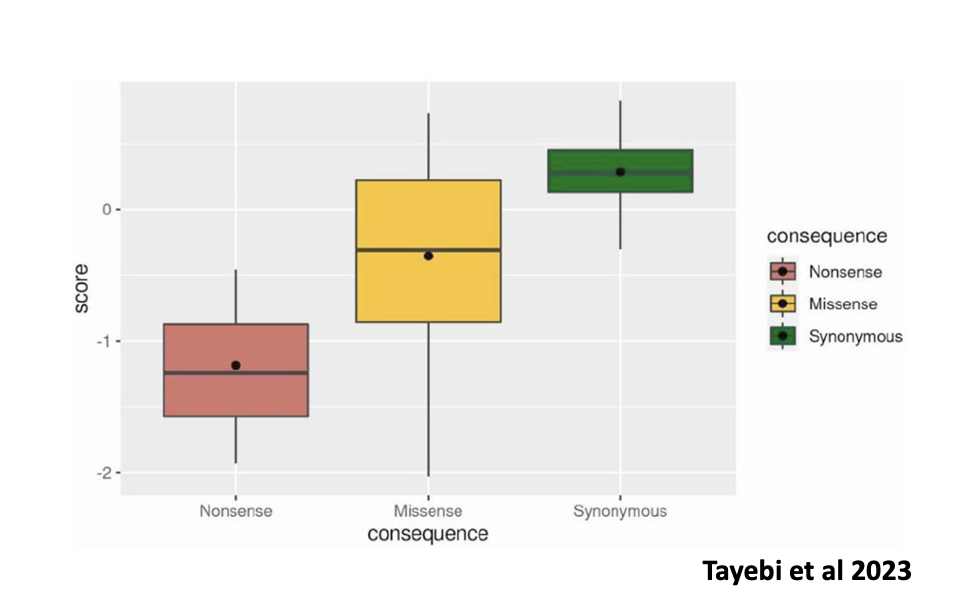
Functional scores of 40 SNVs derived from HAP1 cells growth assay.
Nonsense (# of samples=3), missense (# of samples=27), and synonymous (# of samples= 10)
The graph above shows the functional scores for all the variants made. The functional assay data was compared to several computational metrics that are currently used to determine a variant type (nonsense, missense, deletion), and the comparison showed a significant correlation between their results and the computational metrics data.
Additionally, the functional assays were compared to another in vitro and structural studies of the GLUT1 protein. One of these in vitro assays is a glucose uptake assay, which measures the amount of glucose that goes inside a cell. The amount of glucose entering the cell indirectly measures how well the GLUT1 transporter is working or not. The results found in these assays were also comparable to the functional assay results.
In conclusion, the HAP1 growth assay is a powerful technique to evaluate gene variants in any gene of interest. The future goals for this study are to use this growth assay and evaluate all possible variants in the SLC2A1 gene through deep mutational scanning (DMS). DMS refers to the approach where all possible variants in a sequence are created and are functionally assayed. DMS studies reduce the cost of analyzing just one variant, in addition to giving the ability to compare and analyze all the variants at the same time. Furthermore, DMS generates a “look-up table” of all possible variants that could be used by clinicians and which could improve diagnosis and potentially reduce the need for an invasive confirmatory test such as a lumbar puncture to evaluate CSF glucose.
Finally, having access to quantitative functional data is really powerful because it allows us to give make a better interpretation of a genetic diagnosis which can give more information not only to the clinicians ordering the diagnostic test, but also to the parents and patients who receive a diagnosis.
We thank Dr. Tayebi, Dr. Gurnett and the rest of her team for working on this wonderful and meaningful project for our community.



